DEM Coreg is a bound-restricted minimizing problem
from importlib import reload
import pandas as pd
import xdem
import xsnow
from xsnow.godh import load_gdf,best_shift_px,get_dh_by_shift_px_gdf,load_gdf
from xsnow.misc import df_sampling_from_dem
reload(xsnow.misc)
reload(xsnow.godh)
reload(xdem)
reload(xdem.coreg)
from importlib import reload
import xsnow
import xsnow.godh
DEM Coreg is a Bound-restricted Minimizing Problem.
- DEM Coreg is a bound-restricted minimizing problem, but suffering from noise.
# Loading the dataset.
# 1 ICESat-2 snow free measurements.
sf_gdf = pd.read_csv('snow_free_land_gdf_normal_qc.csv', sep='\t', encoding='utf-8')
# 2 DTM 10 exmaple (10 m Norway DTM-10)
#fid_dtm10 = r'\\hypatia.uio.no\lh-mn-geofag-felles\projects\snowdepth\zhihaol\data\dtm10_finse_merged.tif'
fid_dtm10 = r'\\hypatia.uio.no\lh-mn-geofag-felles\projects\snowdepth\data\DEM\Norway\DTM10_UTM33\6702_4_10m_z33.tif'
dtm_10_ = xdem.DEM(fid_dtm10)
dtm_10 = dtm_10_ + xdem.DEM('no_kv_HREF2018B_NN2000_EUREF89.tif').reproject(dtm_10_,resampling='bilinear')
# 3 DTM 1 exmaple (1 m Norway DTM-1)
fid_dtm1 = r'\\hypatia.uio.no\lh-mn-geofag-felles\projects\snowdepth\data\DEM\Norway\DTM1_UTM33\DTM1_11-11_UTM33\33-113-119.tif'
dtm_1_ = xdem.DEM(fid_dtm1)
dtm_1 = dtm_1_ + xdem.DEM('no_kv_HREF2018B_NN2000_EUREF89.tif').reproject(dtm_1_,resampling='bilinear')
# subset snow free measurements
from xsnow.godh import load_gdf
sf_subset_dtm10_ = load_gdf(sf_gdf,dtm_10,z_name='h_te_best_fit')
sf_subset_dtm1_ = load_gdf(sf_gdf,dtm_1,z_name='h_te_best_fit')
print(f'The length of sf_subset_dtm10: {len(sf_subset_dtm10_)} points')
print(f'The length of sf_subset_dtm1: {len(sf_subset_dtm1_)} points')
The length of sf_subset_dtm10: 44149 points
The length of sf_subset_dtm1: 1586 points
from itertools import product
import numpy as np
import matplotlib.pyplot as plt
import numpy as np
import xdem
from xsnow.misc import df_sampling_from_dem
def yield_surface_noise(dtm_10,sf_gdf_subset,
bond,perc_t=100,std_t=3,stat='nmad', number_corg=None, downsampling_list=None,z_name='h_te_best_fit',
nuthkaab=1,gdc_qc=1,weight=None):
if number_corg is None:
number_corg = 1
if downsampling_list:
for sampling in downsampling_list:
res = [best_shift_px(dtm_10, sf_gdf_subset, disp=False,stat=stat,perc_t=perc_t,std_t=std_t, downsampling=sampling,z_name=z_name,weight=weight) for _ in range(number_corg)]
nmad,rmse,std,median = surface_noise(dtm_10,sf_gdf_subset,bond,perc_t=perc_t,std_t=std_t,downsampling=sampling,z_name=z_name)
if nuthkaab == 1:
try:
func = xdem.coreg.NuthKaab()
func.fit_pts(sf_gdf_subset.sample(frac=sampling/len(sf_gdf_subset),random_state=42),dtm_10,mask_highcurv=False)
res_nk = (func._meta["offset_east_px"],func._meta["offset_north_px"],func._meta["bias"],func._meta["nmad"])
print(f'NuthKaab coreg on {sampling}: {res_nk}')
except ValueError:
res_nk = None
else:
res_nk = None
if gdc_qc == 1:
sf_subset_dtm10_ref = sf_gdf_subset.query('subset_te_flag == 5')
res_qc = best_shift_px(dtm_10, sf_subset_dtm10_ref, disp=False,stat=stat,perc_t=perc_t,std_t=std_t, downsampling=sampling,z_name=z_name)
else:
res_qc = None
yield nmad, rmse, std, median,res,res_nk,res_qc,min(sampling,len(sf_gdf_subset))
def surface_noise(dtm_10,sf_gdf_subset,bond,perc_t=100,std_t=None,downsampling=False,z_name='h_te_best_fit'):
if isinstance(sf_gdf_subset,xdem.DEM):
sf_gdf_subset = df_sampling_from_dem(dtm_10,sf_gdf_subset,samples=10000)
if downsampling and len(sf_gdf_subset) > downsampling:
sf_gdf_subset = sf_gdf_subset.sample(frac=downsampling/len(sf_gdf_subset),random_state=42).copy()
print('Running on downsampling. The length of the gdf:',len(sf_gdf_subset))
print('Set downsampling = other value or False to make a change.')
bond_ = np.arange(bond[0],bond[1],bond[2])
nmad = np.zeros((len(bond_),len(bond_)))
rmse = np.zeros((len(bond_),len(bond_)))
std = np.zeros((len(bond_),len(bond_)))
median = np.zeros((len(bond_),len(bond_)))
for e,n in product(bond_,bond_):
results = get_dh_by_shift_px_gdf(dtm_10,sf_gdf_subset,(e,n),0,stat='all',perc_t=perc_t,std_t=std_t,z_name=z_name)
row = int((n-bond[0])/bond[2])
col = int((e-bond[0])/bond[2])
nmad[row,col] = results['nmad']
rmse[row,col] = results['rmse']
std[row,col] = results['std']
median[row,col] = results['median']
return nmad,rmse,std,median
def surface_plot(Z,bond,xlim=None,ylim=None,zlim=(0,2),
ax=None,title=None,
res=None,res_1=None,res_2=None,res_3=None, minimal=False,zlabel='[m]',
z_step_level=None,zlim_s=20):
if ax is None:
fig, ax = plt.subplots(subplot_kw={"projection": "3d"})
if xlim is None:
xlim=bond[:2]
if ylim is None:
ylim=bond[:2]
# Make a grid.
X = Y = np.arange(bond[0],bond[1],bond[2])
X, Y = np.meshgrid(X, Y)
# plot minimal
zmin = np.min(Z)
zmax = np.max(Z)
mask = np.array(Z) == zmin
#color = np.where(mask,'blue','red')
# Plot the 3D surface
ax.plot_surface(X, Y, Z, edgecolor='royalblue', lw=0.5, rstride=8, cstride=8,
alpha=0.3)
# Plot projections of the contours for each dimension. By choosing offsets
# that match the appropriate axes limits, the projected contours will sit on
# the 'walls' of the graph.
kwargs = {}
if z_step_level:
kwargs['levels'] = np.arange(zmin, zmax + z_step_level, z_step_level)
else:
z_step_level = zlim[1] / zlim_s
kwargs['levels'] = np.arange(zmin, zmax + z_step_level, z_step_level)
z_contour = ax.contour(X, Y, Z,zdir='z', offset=zlim[0], cmap='coolwarm',**kwargs)
ax.contour(X, Y, Z, zdir='x', offset=xlim[0], cmap='coolwarm')
ax.contour(X, Y, Z, zdir='y', offset=ylim[1], cmap='coolwarm_r')
#ax.clabel(z_contour, kwargs['levels'][0::2], z_contour.levels[::2], inline=True,fmt ='%1.1f', colors='black', fontsize='x-large')
if minimal:
# scatter minimal on bottom
ax.scatter(X[mask], Y[mask], zlim[0], color='blue',label='minimal')
#scatter minimal on z-axis
ax.scatter(xlim[1], ylim[1], zmin,marker="x",color='blue')
# plot gradient results
if res:
# plot [0,0] on bottom
ax.scatter(0, 0, zlim[0],alpha=0.4, color='black',edgecolors='white',label='(0,0)')
# plot [shift matrix] by gradient descending on bottom
_x,_y = np.array(res)[:,0],np.array(res)[:,1]
_nmad,_rmse,_std = np.array(res)[:,3],np.array(res)[:,4],np.array(res)[:,5]
ax.scatter(_x, _y, zlim[0],alpha=0.5, s=7,color='green',label='GDC')
# plot shift matrix on z axis
if zlabel == 'NMAD [m]':
_z = _nmad
if zlabel == 'RMSE [m]':
_z = _rmse
if zlabel == 'STD [m]':
_z = _std
#ax.scatter([xlim[1]]*len(_x), [ylim[1]]*len(_y),_z, marker="x", color='green')
# plot nuthkaab results
if res_1:
# plot [shift matrix] by gradient descending on bottom
ax.scatter(res_1[0], res_1[1], zlim[0],alpha=0.5, s=10,color='red',label='NuthKaab')
# plot reference results
if res_2:
# plot [shift matrix] by gradient descending on bottom
ax.scatter(res_2[0], res_2[1], zlim[0],alpha=1, s=10,color='purple',label='GDC_qc')
if res_3:
# plot [shift matrix] by gradient descending on bottom
ax.scatter(res_2[0], res_2[1], zlim[0],alpha=1, s=10,color='black',label='others')
ax.set(xlim=xlim, ylim=ylim, zlim=zlim,
xlabel='Shifted E [px]', ylabel='Shifted N [px]', zlabel=zlabel,title=title)
ax.legend()
Case 1 - Shifted DEM
Nuth & Kaab (2011) showed elevation difference in a 2D scheme with $\alpha$ as the magnitude of the horizontal shift, b as the direction of the shift, which is extremely useful for coregistration when there are enough points and the size and resolution of DEM are relatively manageable.

We can also plot the variation of elevation difference in a 3D scheme. In this case, I shifted a DTM10 (1.2, -0.7) px horizontaly and 0.16 m vertically on purpose and co-register it back in a brute test.
- Brute test calculated the elevation difference in 0.1 px interval over a window (-2 px, 2 px) on east and north.
- All coregs are shifted base on NMAD.
- The points are subsampled to 10000, 3000, 1000.
from xsnow.misc import dem_shift
# Shift a DTM10 (1.2,-0.7) px and 0.16 m on purpose
dtm_10_shifted = dem_shift(dtm_10,1.2,-0.7,0.16)
dem_tba = dtm_10_shifted
df_ref = df_sampling_from_dem(dtm_10,dem_tba,samples=20000)
bond = (-2,2,0.1)
xlim=(-2,2)
ylim=(-2,2)
for nmad, rmse, std, median,res,res_nk,res_qc,sampling in yield_surface_noise(dem_tba,df_ref,bond=bond,perc_t=100,std_t=3,downsampling_list=[10000,3000,1000],gdc_qc=0,z_name='z'):
z_lim_nmad = (max(0,res[0][3]-1),res[0][3]*20)
z_lim_rmse = (max(0,res[0][4]-1.5),res[0][4]*20-1)
z_lim_std = (max(0,res[0][5]-1.5),res[0][5]*20-1)
fig,(ax1,ax2,ax3) = plt.subplots(1,3, figsize=(18, 5),subplot_kw={"projection": "3d"})
fig.suptitle(f'shifted pixels of DEM longyearbyen on {sampling} points', fontsize=16)
surface_plot(nmad, bond=bond,xlim=xlim,ylim=ylim,ax=ax1,title='NMAD', res=res,res_1=res_nk, res_2=res_qc, zlabel='NMAD [m]',zlim=z_lim_nmad,zlim_s=20)
surface_plot(rmse,bond=bond,xlim=xlim,ylim=ylim,ax=ax2,title='RMSE',res=res,res_1=res_nk,res_2=res_qc, zlabel='RMSE [m]',zlim=z_lim_rmse,zlim_s=20)
surface_plot(std,bond=bond,xlim=xlim,ylim=ylim,ax=ax3,title='STD',res=res,res_1=res_nk,res_2=res_qc, zlabel='STD [m]',zlim=z_lim_std,zlim_s=20)
plt.show()
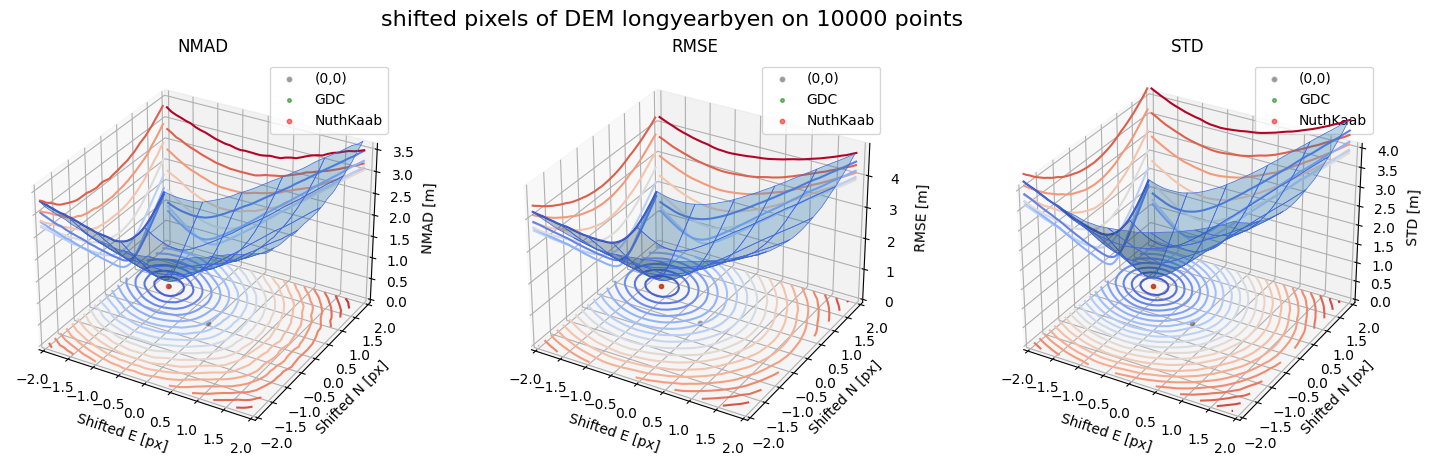
Running best_shift_px on downsampling. The length of the gdf: 10000
Gradient Descending Coreg fit matrix(e_px,n_px,bias),nmad:(-1.2031,0.7031,-0.1630),0.1780
Running on downsampling. The length of the gdf: 10000
Set downsampling = other value or False to make a change.
Warning: There is no curvature in dataframe. Set mask_highcurv = True for more robust results and costs more time
NuthKaab coreg on 10000: (-1.2059757056614222, 0.6973771900864857, -0.16427612, 0.17899064941406248)
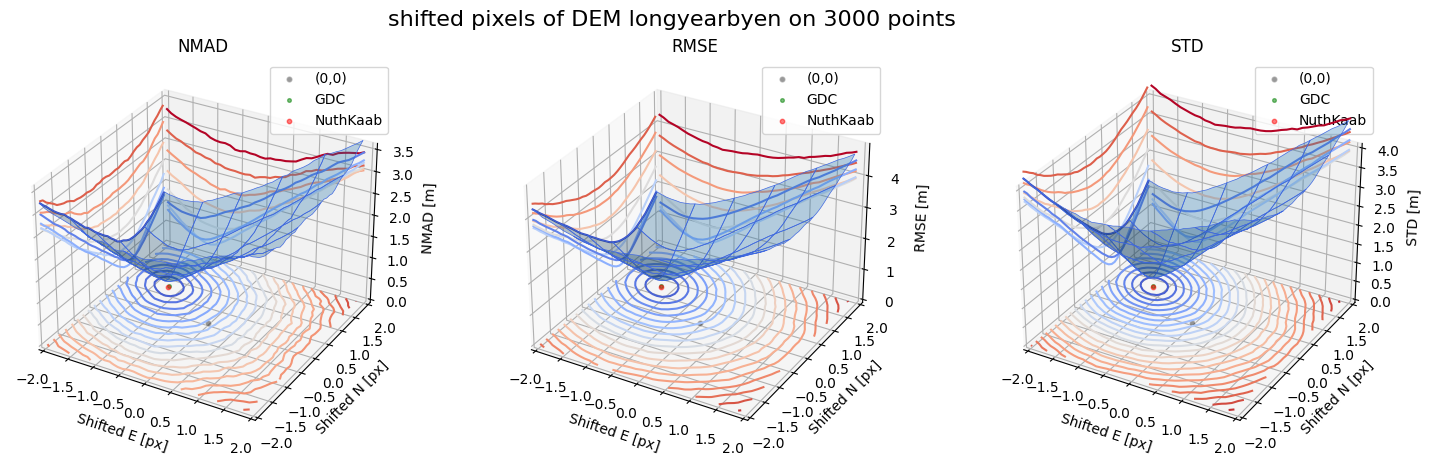
Running best_shift_px on downsampling. The length of the gdf: 3000
Gradient Descending Coreg fit matrix(e_px,n_px,bias),nmad:(-1.2031,0.7031,-0.1630),0.1780
Running on downsampling. The length of the gdf: 3000
Set downsampling = other value or False to make a change.
Warning: There is no curvature in dataframe. Set mask_highcurv = True for more robust results and costs more time
NuthKaab coreg on 3000: (-1.199313462032361, 0.6898301972399311, -0.16362, 0.1797598205566406)
Running best_shift_px on downsampling. The length of the gdf: 1000
Gradient Descending Coreg fit matrix(e_px,n_px,bias),nmad:(-1.1815,0.6815,-0.1622),0.1794
Running on downsampling. The length of the gdf: 1000
Set downsampling = other value or False to make a change.
Warning: There is no curvature in dataframe. Set mask_highcurv = True for more robust results and costs more time
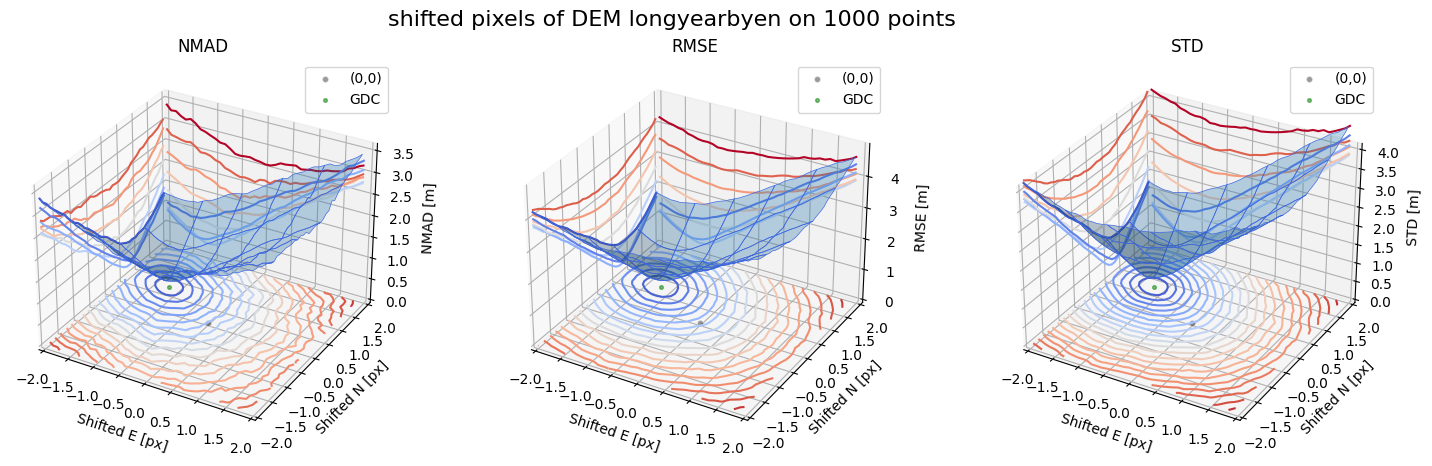
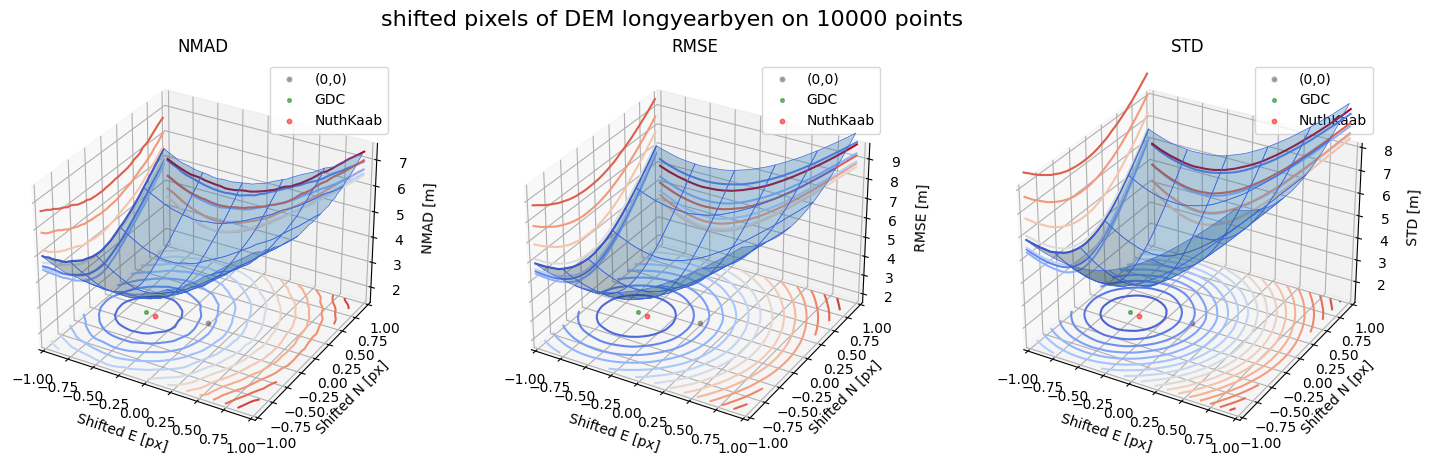
Case 2 - DEM Longyearbyen 1990 and Longyearbyen 2009
from xsnow.misc import df_sampling_from_dem
from xdem import examples
from geoutils import Vector,Raster
dem_ref = xdem.DEM(examples.get_path("longyearbyen_ref_dem"))
dem_tba = xdem.DEM(examples.get_path("longyearbyen_tba_dem"))
glacier_mask = Vector(examples.get_path("longyearbyen_glacier_outlines"))
inlier_mask = ~glacier_mask.create_mask(dem_ref)
df_ref = df_sampling_from_dem(dem_ref,dem_tba,inlier_mask=inlier_mask,samples=15000, offset='ul')
bond = (-1,1,0.05)
# axes limit
xlim=(-1,1)
ylim=(-1,1)
for nmad, rmse, std, median,res,res_nk,res_qc,sampling in yield_surface_noise(dem_tba, df_ref,bond=bond,perc_t=100,std_t=3,downsampling_list=[10000,3000,1000],gdc_qc=0,z_name='z'):
z_lim_nmad = (max(0,res[0][3]-1),res[0][3]*3)
z_lim_rmse = (max(0,res[0][4]-1.5),res[0][4]*3)
z_lim_std = (max(0,res[0][5]-1.5),res[0][5]*3)
fig,(ax1,ax2,ax3) = plt.subplots(1,3, figsize=(18, 5),subplot_kw={"projection": "3d"})
fig.suptitle(f'shifted pixels of DEM longyearbyen on {sampling} points', fontsize=16)
surface_plot(nmad, bond=bond,xlim=xlim,ylim=ylim,ax=ax1,title='NMAD', res=res,res_1=res_nk, res_2=res_qc, zlabel='NMAD [m]',zlim=z_lim_nmad,zlim_s=20)
surface_plot(rmse,bond=bond,xlim=xlim,ylim=ylim,ax=ax2,title='RMSE',res=res,res_1=res_nk,res_2=res_qc, zlabel='RMSE [m]',zlim=z_lim_rmse,zlim_s=20)
surface_plot(std,bond=bond,xlim=xlim,ylim=ylim,ax=ax3,title='STD',res=res,res_1=res_nk,res_2=res_qc, zlabel='STD [m]',zlim=z_lim_std,zlim_s=20)
plt.show()
Running best_shift_px on downsampling. The length of the gdf: 10000
Gradient Descending Coreg fit matrix(e_px,n_px,bias),nmad:(-0.5469,-0.1250,-1.8884),2.4996
Running on downsampling. The length of the gdf: 10000
Set downsampling = other value or False to make a change.
Warning: There is no curvature in dataframe. Set mask_highcurv = True for more robust results and costs more time
NuthKaab coreg on 10000: (-0.44989970346639757, -0.13631736964576785, -1.9579468, 2.543117335510254)
Running best_shift_px on downsampling. The length of the gdf: 3000
Gradient Descending Coreg fit matrix(e_px,n_px,bias),nmad:(-0.5000,-0.1562,-1.8764),2.5242
Running on downsampling. The length of the gdf: 3000
Set downsampling = other value or False to make a change.
Warning: There is no curvature in dataframe. Set mask_highcurv = True for more robust results and costs more time
NuthKaab coreg on 3000: (-0.43152742135357697, -0.17112353846215173, -1.9326782, 2.547257286071777)
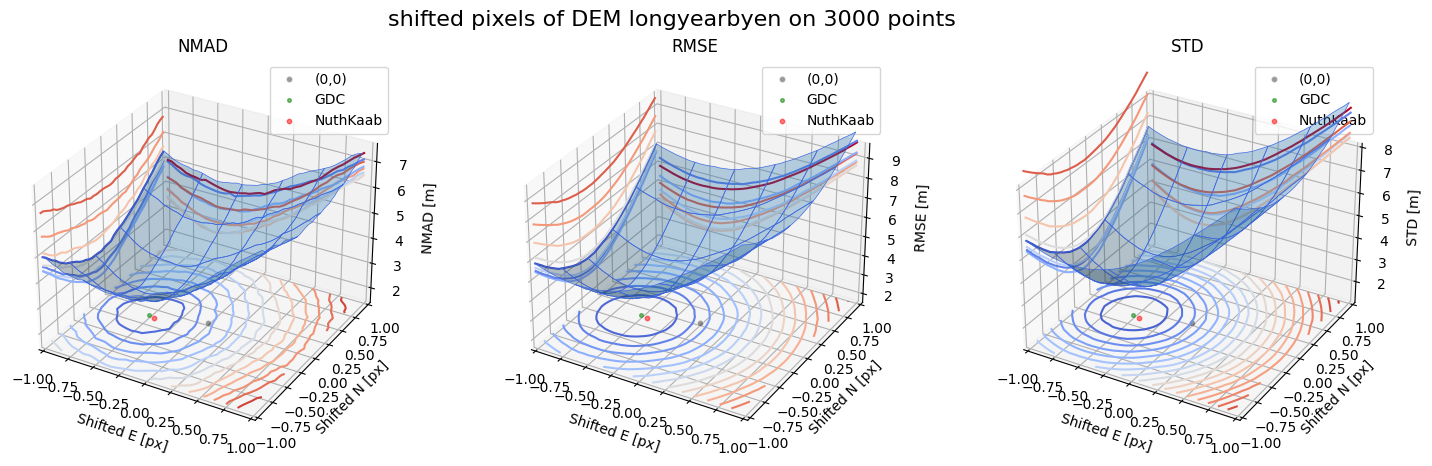
Running best_shift_px on downsampling. The length of the gdf: 1000
Gradient Descending Coreg fit matrix(e_px,n_px,bias),nmad:(-0.5000,-0.1250,-1.8980),2.5237
Running on downsampling. The length of the gdf: 1000
Set downsampling = other value or False to make a change.
Warning: There is no curvature in dataframe. Set mask_highcurv = True for more robust results and costs more time
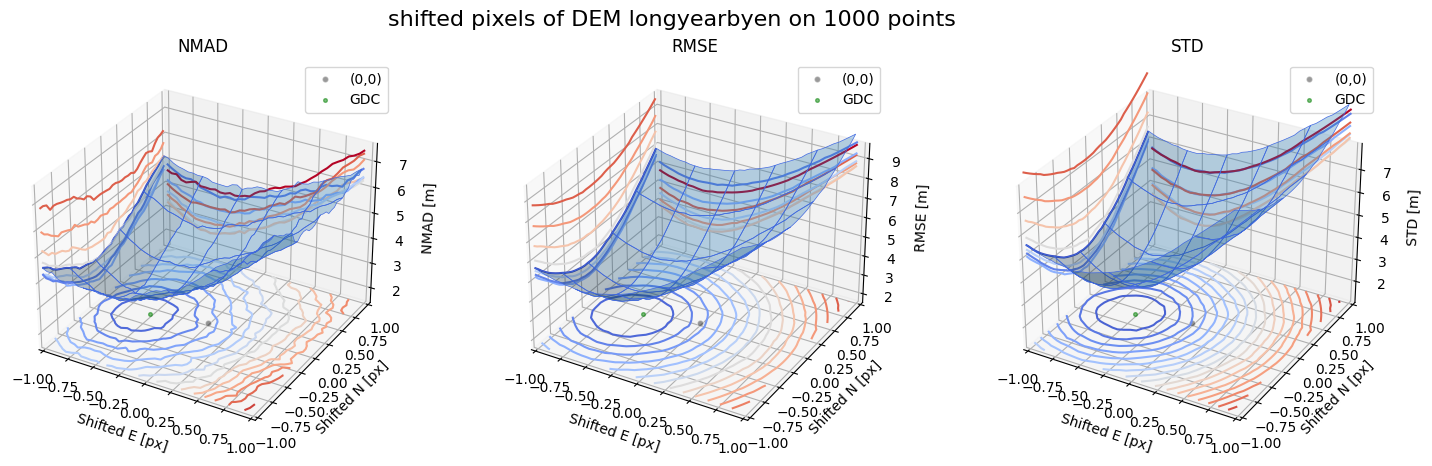
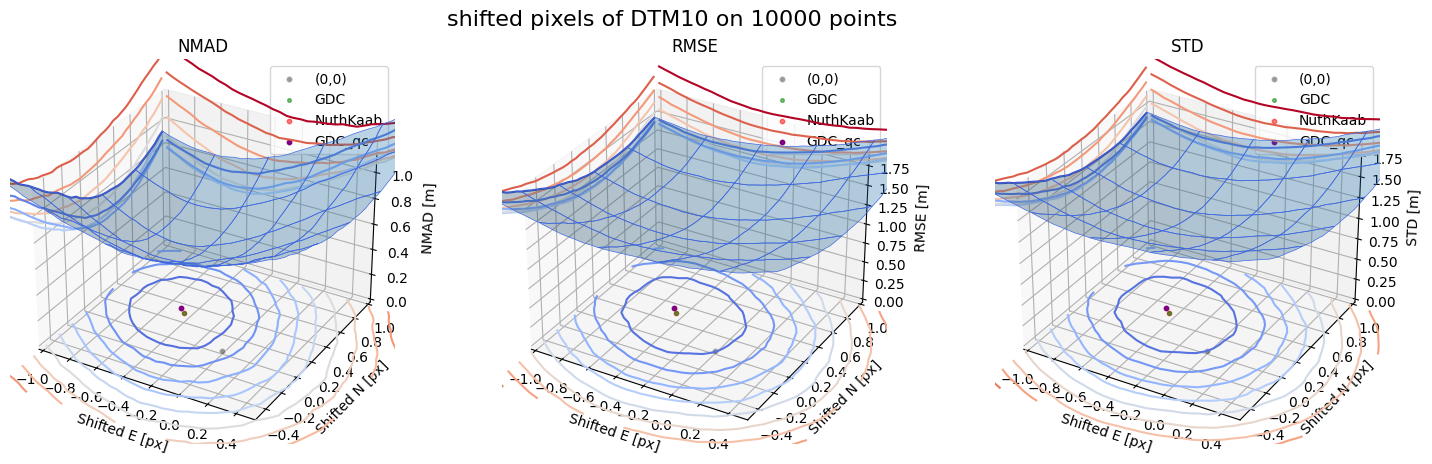
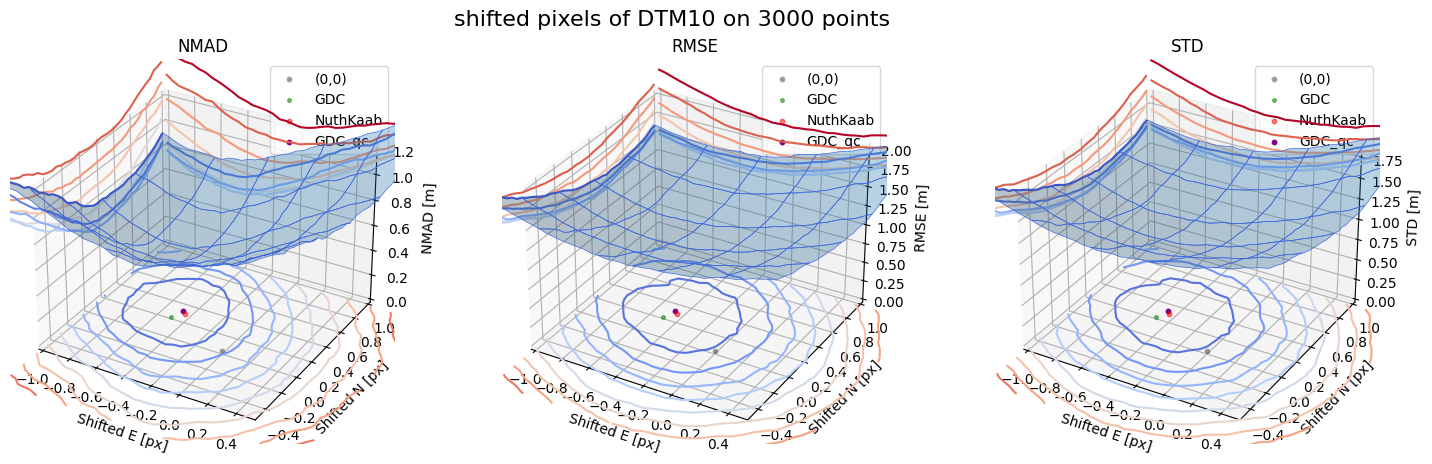
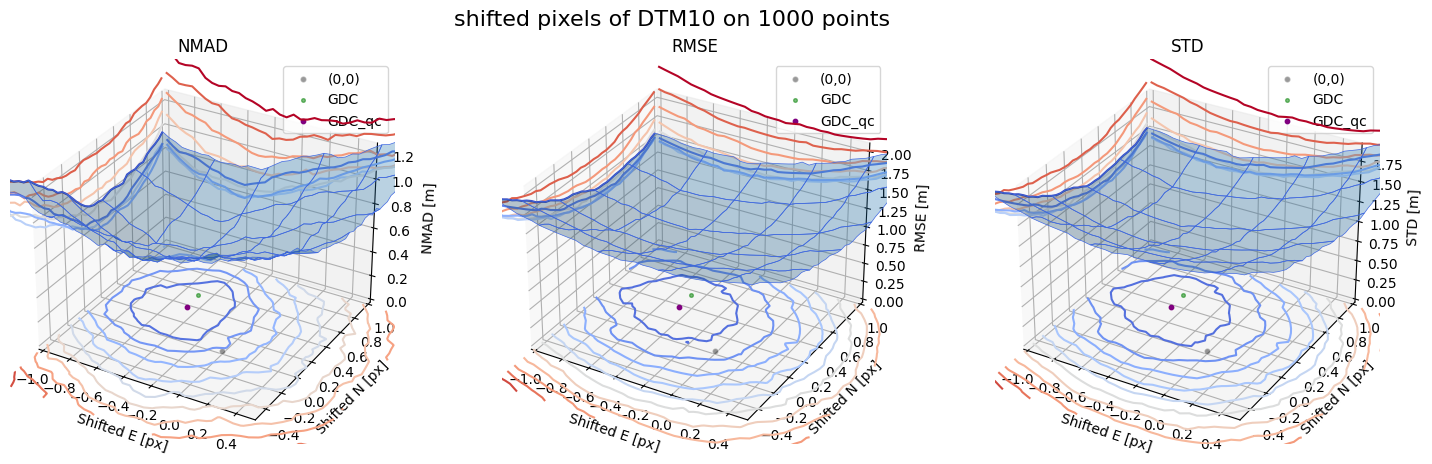
Case 3 - DTM10
bond = (-1.0,1.0,0.05)
# axes limit
xlim=(-1.0,0.5)
ylim=(-0.5,1.0)
for nmad, rmse, std, median,res,res_nk,res_qc,sampling in yield_surface_noise(dtm_10,sf_subset_dtm10,bond=bond,perc_t=100,std_t=3,downsampling_list=[10000,3000,1000],weight='subset_te_flag'):
z_lim_nmad = (0,res[0][3]*2)
z_lim_rmse = (0,res[0][4]*2-1)
z_lim_std = (0,res[0][5]*2-1)
fig,(ax1,ax2,ax3) = plt.subplots(1,3, figsize=(18, 5),subplot_kw={"projection": "3d"})
fig.suptitle(f'shifted pixels of DTM10 on {sampling} points', fontsize=16)
surface_plot(nmad, bond=bond,xlim=xlim,ylim=ylim,ax=ax1,title='NMAD', res=res,res_1=res_nk, res_2=res_qc, zlabel='NMAD [m]',zlim=z_lim_nmad,z_step_level=0.1)
surface_plot(rmse,bond=bond,xlim=xlim,ylim=ylim,ax=ax2,title='RMSE',res=res,res_1=res_nk,res_2=res_qc, zlabel='RMSE [m]',zlim=z_lim_rmse,z_step_level=0.1)
surface_plot(std,bond=bond,xlim=xlim,ylim=ylim,ax=ax3,title='STD',res=res,res_1=res_nk,res_2=res_qc, zlabel='STD [m]',zlim=z_lim_std,z_step_level=0.1)
plt.show()
Running best_shift_px on downsampling. The length of the gdf: 10000
Gradient Descending Coreg fit matrix(e_px,n_px,bias),nmad:(-0.4375,0.2656,-0.0065),0.5992
Running on downsampling. The length of the gdf: 10000
Set downsampling = other value or False to make a change.
Warning: There is no curvature in dataframe. Set mask_highcurv = True for more robust results and costs more time
NuthKaab coreg on 10000: (-0.43959885000839216, 0.2685718552192047, -0.0128173828125, 0.5952479736328125)
Running best_shift_px on downsampling. The length of the gdf: 10000
Gradient Descending Coreg fit matrix(e_px,n_px,bias),nmad:(-0.4844,0.3125,0.1088),0.3848
Running best_shift_px on downsampling. The length of the gdf: 3000
Gradient Descending Coreg fit matrix(e_px,n_px,bias),nmad:(-0.4844,0.1815,-0.0063),0.6086
Running on downsampling. The length of the gdf: 3000
Set downsampling = other value or False to make a change.
Warning: There is no curvature in dataframe. Set mask_highcurv = True for more robust results and costs more time
NuthKaab coreg on 3000: (-0.4273086350909474, 0.259520889514669, -0.01318359375, 0.5863798828125)
Running best_shift_px on downsampling. The length of the gdf: 3000
Gradient Descending Coreg fit matrix(e_px,n_px,bias),nmad:(-0.4591,0.2909,0.1092),0.3856
Running best_shift_px on downsampling. The length of the gdf: 1000
Gradient Descending Coreg fit matrix(e_px,n_px,bias),nmad:(-0.4688,0.5060,-0.0110),0.6305
Running on downsampling. The length of the gdf: 1000
Set downsampling = other value or False to make a change.
Warning: There is no curvature in dataframe. Set mask_highcurv = True for more robust results and costs more time
Running best_shift_px on downsampling. The length of the gdf: 1000
Gradient Descending Coreg fit matrix(e_px,n_px,bias),nmad:(-0.4627,0.3438,0.1067),0.3884
from xsnow.godh import get_dh_by_shift_px_gdf
get_dh_by_shift_px_gdf(dtm_10,sf_subset_dtm10,(-0.4375,0.2656),perc_t=100,std_t=3)
0.5992295654296874
get_dh_by_shift_px_gdf(dtm_10,sf_subset_dtm10,(-0.43959885000839216, 0.2685718552192047),perc_t=100,std_t=3)
0.5986866210937499
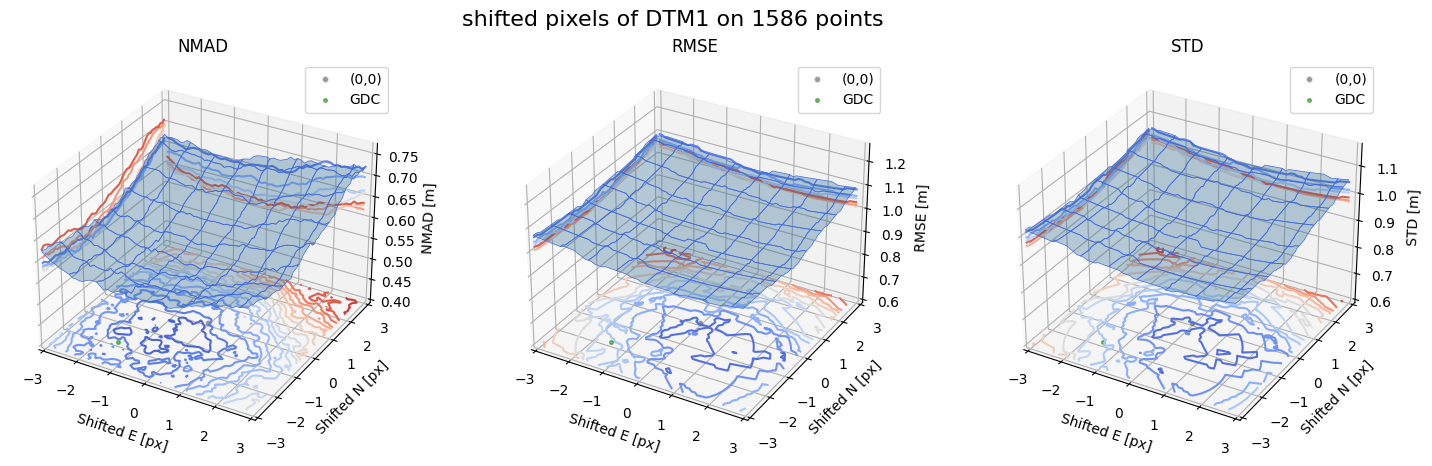
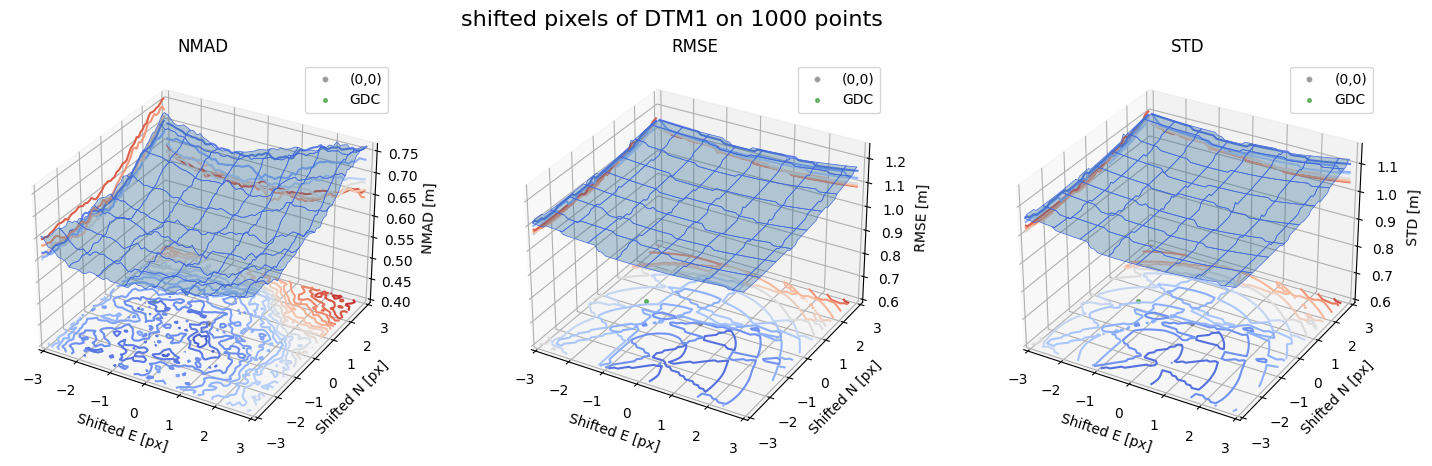
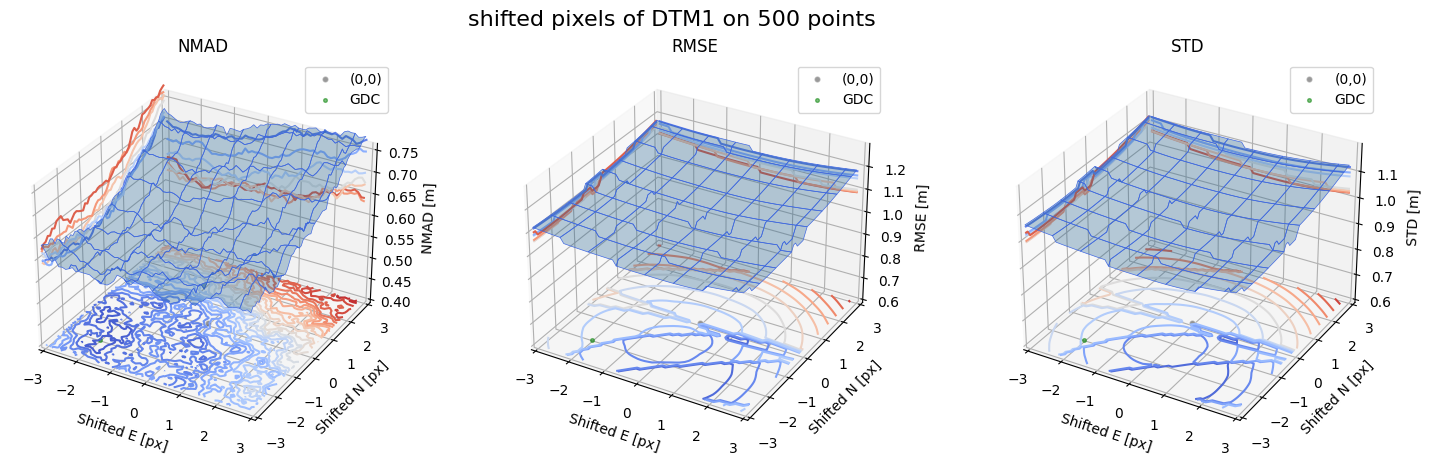
Case - 4 DTM1
bond = (-3,3,0.1)
# axes limit
xlim=(-3,3)
ylim=(-3,3)
for nmad, rmse, std, median,res,res_nk,res_qc,sampling in yield_surface_noise(dtm_1,sf_subset_dtm1,bond=bond,perc_t=100,std_t=3,downsampling_list=[10000,1000,500],gdc_qc=0,weight='subset_te_flag'):
z_lim_nmad = (0.4,res[0][3]*2-0.4)
z_lim_rmse = (0.6,res[0][4]*2-0.8)
z_lim_std = (0.6,res[0][5]*2-0.8)
fig,(ax1,ax2,ax3) = plt.subplots(1,3, figsize=(18, 5),subplot_kw={"projection": "3d"})
fig.suptitle(f'shifted pixels of DTM1 on {sampling} points', fontsize=16)
surface_plot(nmad, bond=bond,xlim=xlim,ylim=ylim,ax=ax1,title='NMAD', res=res,res_1=res_nk, res_2=res_qc, zlabel='NMAD [m]',zlim=z_lim_nmad,z_step_level=0.01)
surface_plot(rmse,bond=bond,xlim=xlim,ylim=ylim,ax=ax2,title='RMSE',res=res,res_1=res_nk,res_2=res_qc, zlabel='RMSE [m]',zlim=z_lim_rmse,z_step_level=0.01)
surface_plot(std,bond=bond,xlim=xlim,ylim=ylim,ax=ax3,title='STD',res=res,res_1=res_nk,res_2=res_qc, zlabel='STD [m]',zlim=z_lim_std,z_step_level=0.01)
plt.show()
Gradient Descending Coreg fit matrix(e_px,n_px,bias),nmad:(-1.5000,-1.9375,-0.0508),0.5833
Running best_shift_px on downsampling. The length of the gdf: 1000
Gradient Descending Coreg fit matrix(e_px,n_px,bias),nmad:(-1.7656,0.2284,-0.0659),0.5798
Running on downsampling. The length of the gdf: 1000
Set downsampling = other value or False to make a change.
Warning: There is no curvature in dataframe. Set mask_highcurv = True for more robust results and costs more time
Running best_shift_px on downsampling. The length of the gdf: 500
Gradient Descending Coreg fit matrix(e_px,n_px,bias),nmad:(-1.9736,-2.0781,-0.0555),0.5788
Running on downsampling. The length of the gdf: 500
Set downsampling = other value or False to make a change.
Warning: There is no curvature in dataframe. Set mask_highcurv = True for more robust results and costs more time
The Best Fit Algorithm and Coefficients
Gradient descending can also be implemented by L-BFGS-B from Scipy.optimize if the noise is not severe.
from scipy.optimize import minimize
from xsnow.godh import get_dh_by_shift_px_gdf
# start iteration, find the best shifting px
func_x = lambda x: get_dh_by_shift_px_gdf(dtm_10,sf_subset_dtm10,x,perc_t=100,std_t=3,stat='rmse')
res = minimize(func_x, x0=(0,0), method='L-BFGS-B',bounds=((-3,3),(-3,3)))
res
fun: 1.5511970187060895
hess_inv: <2x2 LbfgsInvHessProduct with dtype=float64>
jac: array([0. , 0.01073706])
message: 'CONVERGENCE: REL_REDUCTION_OF_F_<=_FACTR*EPSMCH'
nfev: 99
nit: 7
njev: 33
status: 0
success: True
x: array([-0.09557894, 0.00446687])
res = minimize(func_x, x0=(0,0), method='BFGS')
res
fun: 1.5797875171900577
hess_inv: array([[7.02123e-09, 0.00000e+00],
[0.00000e+00, 1.00000e+00]])
jac: array([0.0354276, 0. ])
message: 'Desired error not necessarily achieved due to precision loss.'
nfev: 237
nit: 2
njev: 75
status: 2
success: False
x: array([1.37243807e-07, 0.00000000e+00])
from xsnow.noiseopt import minimizeCompass
res = minimizeCompass(func_x, x0=(0,0), deltainit=2,deltatol=0.006,feps=0.0001,bounds=((-3,3),(-3,3)),errorcontrol=False)
res
free: array([False, False])
fun: 1.491898243075958
message: 'convergence within deltatol'
nfev: 41
nit: 14
success: True
x: array([-0.359375, 0.25 ])
res = minimizeCompass(func_x, x0=(0,0), deltainit=2,deltatol=0.006,feps=0.0005,bounds=((-3,3),(-3,3)),errorcontrol=False)
res
free: array([ True, False])
fun: 1.491898243075958
message: 'convergence within deltatol. dim 0 is free at optimum'
nfev: 40
nit: 13
success: True
x: array([-0.359375, 0.25 ])